Cladistics
19.13
Diposting oleh zakky amarullah
Cladistics (ancient Greek: κλάδος, klados, "branch") is a form of biological systematics that classifies species of organisms into hierarchical monophyletic groups. It can be distinguished from other taxonomic systems, such as phenetics, by its focus on shared derived characters (synapomorphies). Previous systems usually employed overall morphological similarity to group species into genera, families and other higher level classification; cladistic classifications (usually trees called cladograms) are intended to reflect the relative recency of common ancestry or the sharing of homologous features. Cladistics is also distinguished by its emphasis on parsimony and hypothesis testing (particularly falsificationism), rather than subjective decisions that some other taxonomic systems rely upon[1].
Cladistics originated in the work of the German entomologist Willi Hennig, who referred to it as "phylogenetic systematics" (also the name of his 1966 book); the use of the terms "cladistics" and "clade" was popularized by other researchers. The technique and sometimes the name have been successfully applied in other disciplines: for example, to determine the relationships between the surviving manuscripts of the Canterbury Tales[2].
Cladists use cladograms, diagrams which show ancestral relations between species, to represent the monophyletic relationships of species, termed sister-group relationships. This is interpreted as representing phylogeny, or evolutionary relationships. Although traditionally such cladograms were generated largely on the basis of morphological characters, genetic sequencing data and computational phylogenetics are now very commonly used in the generation of cladograms.
History of cladistics
The term clade was introduced in 1958 by Julian Huxley, cladistic by Cain and Harrison in 1960, and cladist (for an adherent of Hennig's school) by Mayr in 1965.[3] Hennig referred to his own approach as phylogenetic systematics. From the time of his original formulation until the end of the 1980s cladistics remained a minority approach to classification. However in the 1990s it rapidly became the dominant method of classification in evolutionary biology. Computers made it possible to process large quantities of data about organisms and their characteristics. At about the same time the development of effective polymerase chain reaction techniques made it possible to apply cladistic methods of analysis to biochemical and molecular genetic features of organisms as well as to anatomical ones.[4]
Cladistics as a successor to phenetics
For some decades in the mid to late twentieth century, a commonly used methodology was phenetics ("numerical taxonomy"). This can be seen as a predecessor[5] to some methods of today's cladistics (namely distance matrix methods such as neighbor-joining), but made no attempt to resolve phylogeny, only similarities.
Clades
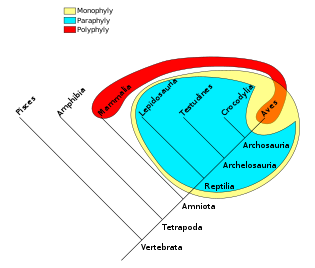
The clade is the topic of interest to cladistics. A clade is a group of taxa consisting of an ancestor taxon and all of its descendant taxa. In the diagram provided, it is hypothesized that the first vertebrate species is the common ancestor of all vertebrate species, including fishes (Pisces). The first tetrapod is the ancestor of all tetrapods, including amphibians, reptiles, mammals and birds. The tetrapod ancestor was a descendant of the original vertebrate ancestor, but is not an ancestor of any fish. The vertebrate clade therefore consists of a fish clade (emcompassing all fish) and a tetrapod clade (consisting of all the tetrapods) and so on up the tree.
An important caution is that any cladogram is a provisional hypothesis. In a hypothetical example, further genetic or morphological evidence might suggest that fish and amphibians share a common ancestor that was not an ancestor of the other tetrapods. The new information would cause us to define a fish-and-amphibian clade, altering the cladogram.
Terminology
The following terms are used to identify shared or distinct characters among groups:[6][7][8]
- Plesiomorphy ("close form") or ancestral state, also symplesiomorphy ("shared plesiomorphy"), is a characteristic that is present at the base of the tree. For example, in the tree shown, the presence of a backbone (shared by all vertebrates) can be hypothesized to have existed in the common vertebrate ancestor.
- Apomorphy ("separate form") or derived state is a characteristic believed to have evolved within the tree. For example, all tetrapods have four limbs; thus, having four limbs is an apomorphy for vertebrates but a plesiomorphy for tetrapods.
- Synapomorphy ("shared apomorphy") is an apomorphy which is shared between taxa.
The application of the above terms to a group depends on one's perspective in the tree. They are thus relative terms. For example, an apomorphy of one clade is the plesiomorphy of another contained within it. These terms are equivalent to but more precise than the homology, allowing one to express the hierarchical relationships among different homologies.
Three main types of groups can be identified by plotting their relationships in cladograms:[6][7][8]
- Monophyletic groups are groups containing only taxa descended from a given ancestor taxon and all those descendants. In the diagram, all vertebrates are monophyletic, since all the taxa are descended from a single ancestor (the common vertebrate species) and there are no others. Monophyly is diagnosed by the synapomorphy relation.
- Paraphyletic groups are groups excluding one or more descendant taxa of the common ancestor, and thus contain some but not all of its descendants. For instance, excluding birds from the sample cladogram would create a paraphyletic group, reptiles. Paraphyletic groups are typically diagnosed on the basis of shared plesiomorphy and the exclusion of groups that are diagnosable by a synapomorphy. For example, birds are warm-blooded, excluding them from being reptiles, which because of that exclusion is a paraphyletic group, as birds are also descended from amniotes. The plesiomorphy is the amniotic fluid and the synapomorphy is warm-bloodedness.
- Polyphyletic groups are groups containing taxa from two or more different monophyletic groups. For instance, a grouping of birds and mammals based on their warm-bloodedness is not monophyletic. In the diagram given (rightly or wrongly) the Aves ancestor came from the Archaeosaurs, but the Mammalian ancestor came from the Amniotes. Although there is an ultimate common ancestor among the Tetrapods, the latest common ancestors are not the same and therefore the group is polyphyletic. The warm-bloodedness is not a plesiomorphy but is a homoplasy, or convergence. Polyphyletic groups are recognized by the homoplasy relation (that is, a group is polyphyletic because it is diagnosed by a character that actually forms a homoplasy, see wastebasket taxon).
Clades relate to each other in these ways:
- A clade is basal to another clade if it contains that other clade as a subset within it. In this case, the vertebrate clade is basal to the tetrapod and fish clades. The use of "basal" to mean a clade that is less species-rich than a sister clade, with such a deficit being taken as an indication of primitiveness, is incorrectly applied.[9]
- A clade located within a clade is said to be nested within that clade. The bird clade is nested within the reptilian clade.
Three definitions of clade
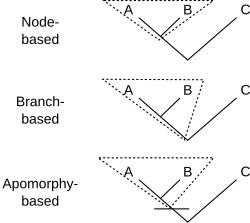
There are three major ways to define a clade for use in a cladistic taxonomy.[10]
- Node-based: the last common ancestor of A and B, and all descendants of that ancestor. Crown groups are a type of node-based clade.
- Branch-based: the first ancestor of A which is not also an ancestor of C, and all descendants of that ancestor. (This type of definition was originally called "stem-based", but this was changed to avoid confusion with the term "stem group".) Total groups are a type of branch-based clade.
- Apomorphy-based: the first ancestor of A to possess derived trait M homologously (that is, synapomorphically) with that trait in A, and all descendants of that ancestor. The process of identifying and naming groups based on apomorphies is the method that most resembles classical systematics, with the proviso that cladistic taxa always denote a clade.
Cladograms
Cladists use cladograms, diagrams which show ancestral relations between taxa, to represent the evolutionary tree of life. Although traditionally such cladograms were generated largely on the basis of morphological characters, molecular sequencing data and computational phylogenetics are now very commonly used in the generation of cladograms.
The starting point of cladistic analysis is a group of species and molecular, morphological, or other data characterizing those species. The end result is a tree-like relationship diagram called a cladogram,[11] or sometimes a dendrogram (Greek for "tree drawing").[12] The cladogram graphically represents a hypothetical evolutionary process. Cladograms are subject to revision as additional data become available.
The terms "evolutionary tree", and sometimes "phylogenetic tree" are often used synonymously with cladogram,[13] but others treat phylogenetic tree as a broader term that includes trees generated with a nonevolutionary emphasis. In cladograms, all species lie at the leaves. The two taxa on either side of a split, with a common ancestor and no additional descendents, are called "sister taxa" or "sister groups".[14] Each subtree, whether it contains only two or a hundred thousand items, is called a "clade". Many cladists require that all forks in a cladogram be 2-way forks. Some cladograms include 3-way or 4-way forks when there are insufficient data to resolve the forking to a higher level of detail (see under phylogenetic tree).
For a given set of species, the number of distinct cladograms that can be drawn (ignoring which cladogram best matches the species characteristics) is:[15]
| Number of species | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | N |
| Number of cladograms | 1 | 3 | 15 | 105 | 945 | 10,395 | 135,135 | 2,027,025 | 34,459,425 | 1*3*5*7*...*(2N-3) |
This superexponential growth of the number of possible cladograms explains why manual creation of cladograms becomes very difficult when the number of species is large. If a cladogram represents N species, the number of levels (the "depth") in the cladogram is on the order of log2(N).[16] For example, if there are 32 species of deer, a cladogram representing deer could be around 5 levels deep (because 25 = 32), although this is really just the lower limit. A cladogram representing the complete tree of life, with about 10 million species, could be about 23 levels deep. This formula gives a lower limit, with the actual depth generally a larger value, because the various branches of the cladogram will not be uniformly deep. Conversely, the depth may be shallower if forks larger than 2-way forks are permitted.
A cladogram tree has an implicit time axis,[17] with time running forward from the base of the tree to the leaves of the tree. If the approximate date (for example, expressed as millions of years ago) of all the evolutionary forks were known, those dates could be captured in the cladogram. Thus, the time axis of the cladogram could be assigned a time scale (e.g. 1 cm = 1 million years), and the forks of the tree could be graphically located along the time axis. Such cladograms are called scaled cladograms. Many cladograms are not of this type, for a variety of reasons:
- They are built from species characteristics that cannot be readily dated (e.g. morphological data in the absence of fossils or other dating information)
- When the characteristic data are DNA/RNA sequences, it is feasible to use sequence differences to establish the relative ages of the forks, but converting those ages into actual years requires a significant approximation of the rate of change[18]
- Even when the dating information is available, positioning the cladogram's forks along the time axis in proportion to their dates may cause the cladogram to become difficult to understand or hard to fit within a human-readable format
Cladistics makes no distinction between extinct and extant species,[19] and it is appropriate to include extinct species in the group of organisms being analyzed. Cladograms that are based on DNA/RNA generally do not include extinct species because DNA/RNA samples from extinct species are rare. Cladograms based on morphology, especially morphological characteristics that are preserved in fossils, are more likely to include extinct species.
Cladistics in taxonomy
Phylogenetic nomenclature contrasted with traditional taxonomy
Most taxonomists have used the traditional approaches of Linnaean taxonomy and later Evolutionary taxonomy to organize life forms. These approaches use several fixed levels of a hierarchy, such as kingdom, phylum, class, order, and family. Phylogenetic nomenclature does not feature those terms, because the evolutionary tree is so deep and so complex that it is inadvisable to set a fixed number of levels.
Evolutionary taxonomy insists that groups reflect phylogenies. In contrast, Linnaean taxonomy allows both monophyletic and paraphyletic groups as taxa. Since the early 20th century, Linnaean taxonomists have generally attempted to make at least family- and lower-level taxa (i.e. those regulated by the codes of nomenclature) monophyletic. Ernst Mayr in 1985 drew a distinction between the terms cladistics and phylogeny:[21]
"It would seem to me to be quite evident that the two concepts of phylogeny (and their role in the construction of classifications) are sufficiently different to require terminological distinction. The term phylogeny should be retained for the broad concept of phylogeny, promoted by Darwin and adopted by most students of phylogeny in the ensuing 90 years. The concept of phylogeny as mere genealogy should be terminologically distinguished as cladistics. To lump the two concepts together terminologically could not help but produce harmful equivocation."
Willi Hennig's pioneering work provoked a spirited debate[22] about the relative merits of phylogenetic nomenclature versus Linnaean or evolutionary taxonomy, which has continued down to the present[23], however Hennig did not advocate abandoning the Linnaean nomenclatural system. Some of the debates in which the cladists were engaged had been running since the 19th century, but they were renewed fervor,[24] as can be seen from the Foreword to Hennig (1979) by Rosen, Nelson, and Patterson:[25]
"Encumbered with vague and slippery ideas about adaptation, fitness, biological species and natural selection, neo-Darwinism (summed up in the "evolutionary" systematics of Mayr and Simpson) not only lacked a definable investigatory method, but came to depend, both for evolutionary interpretation and classification, on consensus or authority."
Phylogenetic nomenclature strictly and exclusively follows phylogeny and has arbitrarily deep trees with binary branching: each taxon corresponds to a clade. Linnaean taxonomy, while since the advent of evolutionary theory following phylogeny, also may subjectively consider similarity and has a fixed hierarchy of taxonomic ranks, and its taxa are not required to correspond to clades.
Paraphyletic groups discouraged
Many cladists discourage the use of paraphyletic groups in classification of organisms, because they detract from cladistics' emphasis on clades (monophyletic groups). In contrast, proponents of the use of paraphyletic groups argue that any dividing line in a cladogram creates both a monophyletic section above and a paraphyletic section below. They also contend that paraphyletic taxa are necessary for classifying earlier sections of the tree – for instance, the early vertebrates that would someday evolve into the family Hominidae cannot be placed in any other monophyletic family. They also argue that paraphyletic taxa provide information about significant changes in organisms' morphology, ecology, or life history – in short, that both paraphyletic groups and clades are valuable notions with separate purposes.
Complexity of the Tree of Life
The cladistic tree of life is a fractal:[26]
"The tree of life is inherently fractal-like in its complexity, .... Look closely at the 'lineage' of a phylogeny ... and it dissolves into many smaller lineages, and so on, sown to a very fine scale."
The overall shape of a dichotomous (bifurcating) tree is recursive; as a viewpoint zooms into the tree of life, the same type of tree appears no matter what the scale. When extinct species are considered (both known and unknown), the complexity and depth of the tree can be very large. Moreover the tree continues to recreate itself by bifurcation, a series of events called fractal evolution.[27]. Every single speciation event, including all the species that are now extinct, represents an additional fork on the hypothetical, complete cladogram of the tree of life.
The tree of life is a quasi-self-similar fractal; that is, the deep reconstruction is not as regular as the shallow reconstruction.[28] By shallow Mishler means the most recent branching toward and at the tips, and by deep the more ancient branches further back, which are harder to reconstruct and are missing unknown extinct lines. In the shallow part of the tree, branching events are relatively regular; it is often possible to estimate the times between them. In the deep part of the tree, "homology assessments" are "difficult" and the times vary widely.[29] At this level Eldredge's and Gould's punctuated equilibrium applies, which hypothesizes long periods of stability followed by punctuations of rapid speciation, based on the fossil record.
PhyloCode approach to naming species
A formal code of phylogenetic nomenclature, the PhyloCode[30], is currently under development. It is intended for use by both those who would like to abandon Linnaean taxonomy and those who would like to use taxa and clades side by side. In several instances (see for example Hesperornithes) it has been employed to clarify uncertainties in Linnaean systematics so that in combination they yield a taxonomy that unambiguously places problematic groups in the evolutionary tree in a way that is consistent with current knowledge.
Example
For example, Linnaean taxonomy contains the taxon Tetrapoda, defined morphologically as vertebrates with four limbs (as well as animals with four-limbed ancestors, such as snakes), which is often given the rank of superclass, and divides into the classes Amphibia, Reptilia, Aves, Mammalia, and some extinct families.
Phylogenetic nomenclature also contains the taxon Tetrapoda (see the diagram under Clades above), whose living members can be classified phylogenically as "the clade defined by the common ancestor of amphibians and mammals", or more precisely the clade defined by the common ancestor of a specific amphibian and mammal (or bird or reptile), but whose tree is still being worked out (there are a number of extinct branches). The taxon does not have a rank, and its subtaxa are subclades: these can be contained within one another, but one does not divide the clade into several non-overlapping taxa (as in traditional taxonomy): one can split into two clades at the first branching, but that is all. With regards to the traditional classes, Aves and Mammalia are subclades, contained in the subclade Amniota, but Reptilia* is a paraphyletic taxon, not a clade — "At best, the cladists suggest, we could say that the traditional Reptilia are 'non-avian, non-mammalian amniotes'"[31] — and instead one divides Amniota into the two clades Sauropsida (which contains birds and all living amniotes other than mammals, including all living traditional reptiles) and Theropsida (mammals and the extinct "mammal-like reptiles"). Similarly, Amphibia* is a paraphyletic taxon.
Summary of advantages of phylogenetic nomenclature
Proponents of phylogenetic nomenclature enumerate key distinctions between phylogenetic nomenclature and Linnaean taxonomy as follows:[32]
| Phylogenetic Nomenclature | Linnaean Taxonomy |
| Handles arbitrarily deep trees. | Often must invent new level names (such as superorder, suborder, infraorder, parvorder, magnorder) to accommodate new discoveries. Biased towards trees about 4 to 12 levels deep. |
| Discourages naming or use of groups that are not monophyletic | Acceptable to name and use paraphyletic groups |
| Primary goal is to reflect actual process of evolution | Primary goal is to group species based on morphological similarities |
| Assumes that the shape of the tree will change frequently, with new discoveries | New discoveries often require renaming or releveling of Classes, Orders, and Kingdoms |
[edit] Summary of criticisms of phylogenetic nomenclature
Critics of phylogenetic nomenclature include Ashlock,[33] Mayr,[34] Williams.[35] Some of their criticisms include:
| Phylogenetic Nomenclature | Linnaean Taxonomy |
| Limited to entities related by evolution or ancestry | Supports groupings without reference to evolution or ancestry |
| Does not include a process for naming species | Includes a process for giving unique names to species |
| Difficult to understand the essence of a clade, because clade definitions emphasize ancestry at the expense of meaningful characteristics | Taxa definitions based on tangible characteristics |
| Ignores sensible, clearly defined paraphyletic groups such as reptiles | Permits clearly defined groups such as reptiles |
| Difficult to determine if a given species is in a clade or not (e.g. if clade X is defined as "most recent common ancestor of A and B along with its descendants", then the only way to determine if species Y is in the clade is to perform a complex evolutionary analysis) | Straightforward process to determine if a given species is in a taxon or not |
| Limited to organisms that evolved by inherited traits; not applicable to organisms that evolved via complex gene sharing or lateral transfer | Applicable to all organisms, regardless of evolutionary mechanism |
[edit] Application to other disciplines
The comparisons used to acquire data on which cladograms can be based are not limited to the field of biology.[36] Any group of individuals or classes, hypothesized to have a common ancestor, and to which a set of common characteristics may or may not apply, can be compared pairwise. Cladograms can be used to depict the hypothetical descent relationships within groups of items in many different academic realms. The only requirement is that the items have characteristics that can be identified and measured.
Recent attempts to use cladistic methods outside of biology address the reconstruction of lineages in:
- Anthropology and archeology.[37] Compares cultures or artifacts using groups of cultural traits or artifact features.
- Linguistics.[38] Compares languages using groups of linguistic features.
- Textual criticism or Stemmatics.[39][40] Compares manuscripts of the same work (original lost) using groups of distinctive copying errors.
- Ethology.[41] Compares animal species using behavioral traits presumed hereditary.
Posting Komentar